FAIR and its origins
Overview
Teaching: 20 min
Exercises: 20 minQuestions
What is FAIR and the FAIR Guiding Principles?
Where does FAIR come from?
Objectives
Identify the FAIR principles and their origin
Explain the difference between FAIR and open data
Contextualise the main principles of FAIR around the common characteristics of identifiers, access, metadata and registration
FAIR
The concepts underlying the FAIR principles are intuitively grounded in good scientific practice, though formalising them as easily measurable is not straightforward. Being completely ‘FAIR’ is somewhat of an ideal, where it is often more pragmatic to be ’FAIR enough’ for a particular purpose or use case.
FAIR’s primary goal is to maximise data reuse by researchers. The FAIR principles enable reuse by helping researchers share and manage their data, though FAIR is not limited to data alone and can be applied to services, software, training and workflows. The word FAIR is an acronym, derived from its major components: ‘F’indable, ‘A’ccessible, ‘I’nteroperable, and ‘R’eusable which form the foundation of the FAIR Guiding Principles.
Findable means that data and its metadata can be found/discovered by humans and computers. Part of this is making rich metadata and keywords available to search engines and data repositories, so that companion data can be discovered.
Accessible means that once discovered, data and metadata can be accessed/downloaded by humans and computers. Typically this means the commitment of the resource to its long term hosting and availability, with a suitable licence, and in appropriate format.
Interoperable means that data and metadata are supplied in formats that can be easily used and interpreted by humans and computers. The file formats and terms (vocabularies) used can be integrated easily with other datasets and software.
Reusable means that metadata is rich, enabling appropriate reuse. Commonly FAIR will encourage the use of community standards for data curation.
These 4 major components form headings for the 15 FAIR Guiding Principles shown in Table 1.1
| The FAIR Guiding Principles | |
|---|---|
| To be Findable: | F1. (meta)data are assigned a globally unique and persistent identifier F2. data are described with rich metadata (defined by R1 below) F3. metadata clearly and explicitly include the identifier of the data it describes F4. (meta)data are registered or indexed in a searchable resource |
| To be Accessible: | A1. (meta)data are retrievable by their identifier using a standardized communications protocol A1.1 the protocol is open, free, and universally implementable A1.2 the protocol allows for an authentication and authorization procedure, where necessary A2. metadata are accessible, even when the data are no longer available |
| To be Interoperable: | I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation. I2. (meta)data use vocabularies that follow FAIR principles I3. (meta)data include qualified references to other (meta)data |
| To be Reusable: | R1. meta(data) are richly described with a plurality of accurate and relevant attributes R1.1. (meta)data are released with a clear and accessible data usage license R1.2. (meta)data are associated with detailed provenance R1.3. (meta)data meet domain-relevant community standards |
Table 1.1: The FAIR guiding principles as described in Wilkinson, M., Dumontier, M., Aalbersberg, I. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 (2016). https://doi.org/10.1038/sdata.2016.18
Exercise
Look at the wording of the FAIR principles in Table 1.1. Which terms are used more than once? Which terms are you seeing for the first time?
Solution
Shared terms are: data (F1, F2, F4, A1, A2, I1, I2, I3, R1, R1.1, R1.2, R1,3), metadata (F1, F2, F3, F4, A1, A2, I1, I2, I3, R1, R1.1, R1.2, R1,3), identifier (F1, F3, A1), protocol (A1, A1.1, A1.2).
Not so obviously shared terms are: vocabularies (I1, I2), access (A1, A1.1, A1.2, R1.1)
Terms which you may be seeing for the first time are: metadata, persistent identifier, searchable resource, standardised communications protocol, authentication and authorisation procedure, knowledge representation, vocabularies, qualified references, data usage license, provenance, domain-relevant community standards.
The aim of this course is to put these principles into context and familiarise learners with terms used commonly around FAIR. A glossary of terms appears at the bottom of this episode as a point of reference.
Open data and FAIR
 Figure 1.1: FAIR is not the same as Open data
Figure 1.1: FAIR is not the same as Open data
Since FAIR promotes data sharing, it is often misunderstood as Open data.
The Open Data handbook defines Open data as “data that can be freely used, reused and redistributed by anyone.” Commonly, FAIR data is open however FAIR compliance does not mandate access without restriction. Instead, for instances where FAIR data is subject to restricted access, the conditions of access need to be stated to be compliant with FAIR, for example access around sensitive data. Often in FAIR data, people adhere to the philosophy of “as open as possible, and as closed as necessary” thereby maximising opportunities to reuse data.
Exercise
Under which circumstances would restricted or closed access to FAIR data be advisable?
Solution
Where data is sensitive or subject to intellectual property. Protecting sensitive data overrules mandating research data should be open access.
Note though that in most cases, people following FAIR principles will be looking to share their data openly. Also note that sensitive data can be released through anonymisation and in many cases subject to controlled access by the authority of the principal investigator or data access committee.
What is meant by FAIRification and FAIRness of data?
FAIRification is the process of making your data FAIR compliant by applying the 15 Guiding Principles shown in Table 1.1. The extent to which you apply these principles defines the FAIRness of your data. In other words, FAIRness refers to the extent by which your data is FAIR and implies some implicit means of measuring its compliance.
FAIR’s origins
A report from the European Commission Expert Group on FAIR data describes the origins of FAIR and its development in 2014-2015 by a FORCE11 Working Group. The following exercise dips into this report and asks you to investigate some of FAIR’s history and foundation.
Exercise
Read page 11 of the European Commission report, under the heading “Origins and definitions of FAIR”. What benefit did the FORCE11 Working Group see to coining the word FAIR?
Solution
The report states: “a FORCE11 Working Group coined the FAIR data definition, latching onto an arresting and rhetorically useful acronym. The wordplay with fairness, in the sense of equity and justice, has also been eloquent in communicating the idea that FAIR data serves the best interests of the research community, and the advancement of science as a public enterprise that benefits society.”
Course structure
During the remainder of this course we will put the FAIR Guiding Principles into context using the 4 FAIR characteristics of metadata, data registration, access and identifiers, and devote an episode to each of these. Whilst we teach, we will map content to the appropriate FAIR principle and define all relevant terms identified.
Glossary of FAIR terms
All terms in this glossary are mentioned in the FAIR Guiding Principles (Table 1.2) and referenced in the following episodes of this course.
| The FAIR Guiding Principles | |
|---|---|
| To be Findable: | F1. (meta)data are assigned a globally unique and persistent identifier F2. data are described with rich metadata (defined by R1 below) F3. metadata clearly and explicitly include the identifier of the data it describes F4. (meta)data are registered or indexed in a searchable resource |
| To be Accessible: | A1. (meta)data are retrievable by their identifier using a standardized communications protocol A1.1 the protocol is open, free, and universally implementable A1.2 the protocol allows for an authentication and authorization procedure, where necessary A2. metadata are accessible, even when the data are no longer available |
| To be Interoperable: | I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation. I2. (meta)data use vocabularies that follow FAIR principles I3. (meta)data include qualified references to other (meta)data |
| To be Reusable: | R1. meta(data) are richly described with a plurality of accurate and relevant attributes R1.1. (meta)data are released with a clear and accessible data usage license R1.2. (meta)data are associated with detailed provenance R1.3. (meta)data meet domain-relevant community standards |
Table 1.2: Terms used by the FAIR Principles that appear in this glossary are highlighted in black.
Globally unique and persistent identifier: a reference to a digital resource such as a dataset, a document, a database, etc, usually given as a URL, that takes the user to that resource. The persistent identifier (PID) is unique, globally, in the sense that it is not used to identify any other digital resource. The PID is persistent, in that it enables a resource to be located long term, preferably permanently. For more see: How to FAIR and FAIR Cookbook, episode 5 (identifiers).
Indexed in a searchable resource: repositories and catalogues that can be queried (search box) are examples of searchable resources. Indexing refers to processes within the architecture of the data repository where (meta)data are organised so that they can be queried based on defined fields. Indexing by an internet search engine (for example, Google) is another example of an indexed and searchable resource. For more see: GO FAIR.
Standardised communications protocol: a method that connects two computers and ensures secure data transfer. Examples of this include the hypertext transfer protocol (http(s)) and the file transfer protocol (ftp) that permit data to be requested and downloaded by selecting a link on a webpage, or launched from within a script, for example using an (application programming interface) API. For more see the Australian Research Data Commons.
Universally implementable: in the context of a standardised communications protocol such as http(s), universally implementable means it can be used by a number of resources and software, for example Firefox, Chrome and Unix commands such as wget. For more see the Australian Research Data Commons.
Authentication and authorisation procedure: data repositories with authentication and authorisation procedures generally require a login (and password) to access (meta)data. For more see the Australian Research Data Commons.
Language for knowledge representation: in the context of (meta)data exchange between computers, (meta)data should be in formats that are universally recognised (interoperable standards). For more see GO FAIR and FAIR Cookbook.
Vocabularies: (or controlled vocabulary) is a dictionary of terms you can use when producing (meta)data. Controlled vocabularies are often shared between databases and communities so by using them you can allow data from different sources to be merged (interoperable), based on a shared understanding of the concepts. Ontologies are related to vocabularies, where terms in the vocabulary are organised by relations between them. A commonly used ontology is the NCBI taxonomy where the term ‘Homo sapiens’ belongs to a hierarchy of parent terms such as ‘Primates’ and ‘Mammalia’. The ontology defines the vocabularies and the parent/child relationships. For more see Ten simple rules for making a vocabulary FAIR.
Qualified references: are terms used to describe relationships to pieces of (meta)data. For more see GO FAIR.
Data usage license: describes the legal rights on how others use your data. For more details on considerations in selecting a licence, or to find out more about types of licence that are available, see RDMkit.
Data provenance: refers to metadata describing the origin of a piece of data, including information such as version, original location of the data, and usually an audit trail up to the current version. For more see RDMkit.
Community standards: are standard guidelines used to structure and exchange data, usually supported by community-developed resources and/or software. In the context of (meta)data, community standards relate often to the standardised ontologies used by a domain of research, and minimum information guidelines allowing data to be interoperable. For more see FAIRDOM.
Machine-readable: though this term is not referenced in the FAIR principles, it is often discussed within the FAIR context. Machine-readable (meta)data is supplied in a structured format that can be read by a computer. For more see Open Data Handbook and RDMkit.
(Meta)data: is shorthand for ‘metadata and data’.
Useful Resources
- The published FAIR Guiding Principles: Wilkinson, M., Dumontier, M., Aalbersberg, I. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 (2016). https://doi.org/10.1038/sdata.2016.18.
- Recipes for data FAIRification, written by domain experts giving real-world examples: FAIR Cookbook
- Documentation and frameworks for data FAIRification. Each of the 15 FAIR principles is put into context with real data examples: GO FAIR
- FAIR walkthrough using examples from across all academic disciplines: How to FAIR
Key Points
FAIR stands for Findable, Accessible, Interoperable and Reusable
Metadata, Identifiers, Registration and Access are 4 key components in the process of FAIRification
FAIR data is as open as possible, and as closed as necessary
Metadata
Overview
Teaching: 20 min
Exercises: 20 minQuestions
What is metadata?
What is good metadata?
Using community standards to write (meta)data
Objectives
Define the term ‘metadata’
Recall examples of community/domain standards that apply to data and metadata
Metadata and the FAIR Principles
Metadata relates most directly to the following 10 FAIR Principles highlighted in Table 2.1. We will discuss and signpost these in this episode.
| The FAIR Guiding Principles | |
|---|---|
| To be Findable: | F1. (meta)data are assigned a globally unique and persistent identifier F2. data are described with rich metadata (defined by R1 below) F3. metadata clearly and explicitly include the identifier of the data it describes F4. (meta)data are registered or indexed in a searchable resource |
| To be Accessible: | A1. (meta)data are retrievable by their identifier using a standardized communications protocol A1.1 the protocol is open, free, and universally implementable A1.2 the protocol allows for an authentication and authorization procedure, where necessary A2. metadata are accessible, even when the data are no longer available |
| To be Interoperable: | I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation I2. (meta)data use vocabularies that follow FAIR principles I3. (meta)data include qualified references to other (meta)data |
| To be Reusable: | R1. meta(data) are richly described with a plurality of accurate and relevant attributes R1.1. (meta)data are released with a clear and accessible data usage license R1.2. (meta)data are associated with detailed provenance R1.3. (meta)data meet domain-relevant community standards |
Table 2.1: The 15 FAIR Guiding Principles. Principles relating directly to metadata are highlighted in black.
What is metadata?
Metadata is information that describes your data - it is data about data.
The provision of ‘rich’ metadata is key to FAIR since it allows data to be found, and enables other researchers to interpret data appropriately. ‘Rich’ in this context refers to extensive metadata, often connecting data to other data or terms (even in other datasets), with qualified references specifying how they are connected.
If a researcher is given access to a dataset (a spreadsheet in CSV format, for example), the data is not usable without meaningful column headings and context. For example, what is this data about? Is it part of a larger dataset and, if so, how is it related? What are the column headings representing, and what are the rows representing? Additionally, if the values in the individual cells are device or assay measurements, what device was used, and what assay?
If you look at the example of the spreadsheet below (Figure 2.1) the metadata are coloured blue and the data are in orange. The data in this example are clinical observations for a cohort of patients and the metadata are the descriptive column headings and additional information about the origin of the data. The column headings provide context for the data values in each cell. The contact information enables a researcher to direct questions about data reuse and context. This raises an interesting point though, because metadata should be rich enough to allow any person to reuse the underlying data without ambiguity (FAIR Principles F2, R1).
Though this Figure 2.1 shows the difference between metadata and data, it does not exemplify the use of rich metadata.
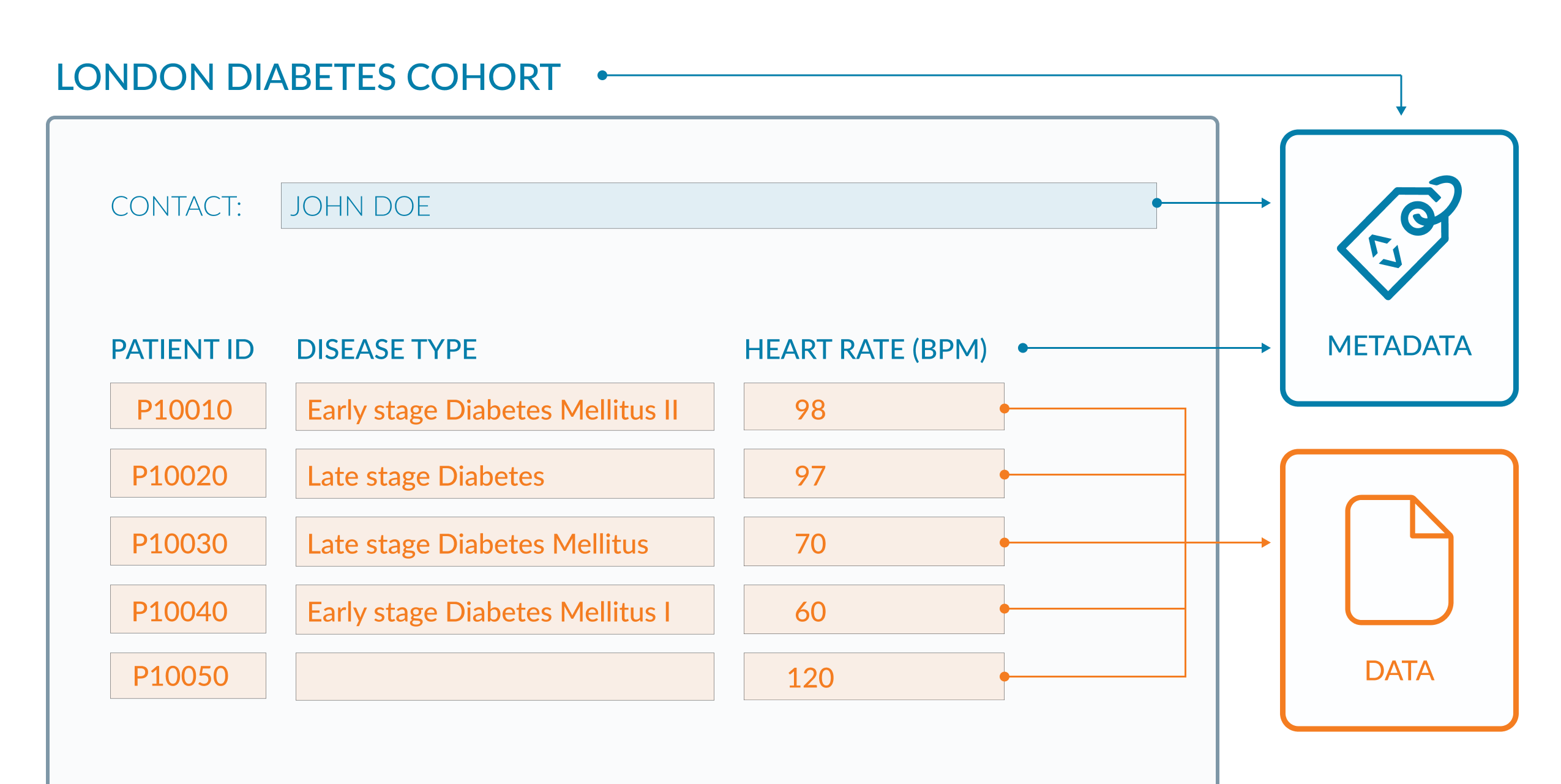
Figure 2.1: A spreadsheet showing the relationship between data (orange) and metadata (blue).
We are given only 2 pieces of data provenance in this example (i) the study name “LONDON DIABETES COHORT” which could help discover other documentation about the project; (ii) the name of the person who presumably led the study. While these may help to search for further information, for a human user, this would still be fraught with issues since both names may not uniquely identify the study or its lead. For a machine agent, such as a script, interpretation is made difficult because the metadata is not clearly marked-up (represented formally with tags).
To fix this, contact details for investigators could be included. Often ORCID IDs associated with names are used since they uniquely identify an individual and do not pose problems when an investigator moves institutions and the original (email) address changes. Coupled with this, investigators can be formally assigned a role in the provenance metadata allowing people to contact the appropriate person, for example, “data producer”, “data manager”.
A project URL should be used where possible, ideally one that can act as a persistent identifier for the dataset. The landing page for that URL should provide more context for the dataset, and describe high level information such as how the dataset relates to other project outputs, as well as providing other provenance information (dates, times, places, people). This could include more metadata about the type of study, objectives, protocols, dates, release versioning and so on. At the end of this episode, resources are given to help define metadata elements that could be included, including a project called Dublin Core which defines 15 such elements or metadata fields.
Terms of access and reuse are missing which could be rectified by including a data licence, which often appears as part of the metadata, usually at the bottom of a webpage hosting data. There could be ambiguity around the acronym, “BPM”, used in the third column header, so this should be defined within a glossary of acronyms and or ideally hyperlinked to a definition in an existing ontology.
There are issues too with the data, as well as the metadata. The second column DISEASE TYPE could be better designed. Two pieces of information (data) are depicted in the same column: disease type (diabetes) and disease stage (early/late). Ideally these should be in 2 separate columns allowing researchers to subset on stage and disease type independently for downstream analysis. There are also four different terms used for diabetes (“Diabetes Mellitus II”, “Diabetes”, “Diabetes Mellitus” and “Diabetes Mellitus I”), which again does not allow a researcher to subset data efficiently. To fix this you would use defined terms within an existing vocabulary or ontology. The following link accesses a disease ontology we could use, where each term (for example, “type 2 diabetes mellitus”) is described and assigned a unique ID. In the example above you would use this unique ID, or the associated descriptive term, to tag all patients with the same disease, identically. This then makes the data sub-setable and machine-readable.
Exercise
Look at the final row of data in Figure 2.1. There is a missing disease type. What problems might this cause?
Solution
This isn’t a metadata problem, but it is worth mentioning that missing data can cause more ambiguity. Does the blank mean the data were not available, or that the data were available but not recorded, or that the individual has no disease? This has implications on how the data are interpreted, and it is likely this data row would be discarded from an analysis. Here you would be looking to use terms to denote missing, for example, “no disease”, “not recorded”.
Writing FAIR metadata
We have discussed already how rich metadata enables a dataset to be reused and interpreted correctly. In the context of the FAIR principles, the previous exercise illustrates two of these, namely that “(Meta)data are richly described with a plurality of accurate and relevant attributes” (FAIR Principle R1) and that “(Meta)data are associated with detailed provenance” (FAIR Principle R1,2). Further to this, the suggested use of the published disease ontology for data, illustrates a further three principles, where “(Meta)data use vocabularies that follow FAIR principles” (FAIR Principle I2), and “(Meta)data meet [domain-relevant community standards] (## “Community standards: standard guidelines used to structure and exchange data, usually supported by community-developed resources and/or software.”)” (FAIR Principle R1.3). The use of hyperlinks specifically to terms in the ontology means that “(Meta)data include qualified referencesto other (meta)data” (FAIR Principle I3). From the previous exercise, the disease ontology provides the vocabulary for the different types of diabetes: type 1 diabetes mellits and type 2 diabetes mellitus.
The FAIR Guiding Principles also highlight the importance of providing rich metadata to enable researchers to find datasets such that “Data are described with rich metadata” (FAIR Principle F2). More often than not, a researcher will find data through searching its metadata, usually via an online or a database search. Information on how this can be achieved is discussed in the next episode on data registration.
The use of vocabularies and cross-references is fundamental to data interoperability. Interoperable (meta)data can be linked and combined across studies, aided by consistent, compatible and machine-readable curation. FAIRsharing is a useful registry of vocabularies, and standards, while more comprehensive ontology lists are maintained by OLS (Ontology Lookup Service) and BioPortal.
The previous exercise also touches on machine-readability of (meta)data through mention of using controlled terms in the “DISEASE TYPE” column to allow subsetting. The Open Data handbook gives a nice overview of machine-readable (meta)data but in short it is (meta)data supplied in a defined and structured format that can easily be read by an appropriate script or piece of software. If we use our example of a spreadsheet in comma-separated value form (CSV format), the (meta)data will be organised into cells, in a format that is interoperable with many software . This would not be true if the same data were made available as a screenshot, highlighting that human-readable data may not be machine-readable.
Rich metadata in public data repositories
Help with rich metadata curation is often supported by public data repositories, and data deposition is one way you can improve its level of FAIRness. During submission, metadata is composed and linked, making it understood, accessible and searchable.
The figure below shows a screenshot of a dataset hosted by the BioStudies (Figure 2.2). BioStudies is a public database holding descriptions for biological studies and their (meta)data, and is often used by researchers to provide primary identifiers to supplementary information described in publications.
 housed in BioStudies showing rich metadata](../fig/figure2-2_rnaseq-database.png)
Exercise
Look at the example given in Figure 2.2. What metadata is given?
Solution
Metadata can be categorised into 3 basic types i (i) descriptive (what the data is); (ii) structural (how the data was generated, i.e. provenance); (iii) administrative (data owners, contributors and funders). In this example, all 3 are given.
Administrative metadata: authors and organisations underneath the dataset title, and the information in “Publication”
Structural metadata: “Protocols” and data links on the right hand side of the page
Descriptive metadata: All other metadata on the left-hand side of the page, describing the study type through to “Samples” and “Assays and Data”. Of note here is the use of controlled vocabularies for metadata such as Organism (taxonomy database) and BioStudy-specific terms for fields such as Experimental Factors, Technology. These allow a researcher to search for all experiments in the database around a theme or type: with queries of the type: “get me all Arabidopsis RNAseq data”.
Using community standards for (meta)data
Public databases often serve communities and specific types of data, and may often use community standards for metadata curation. These standards include, usually, open-access ontologies that can be used by researchers to annotate their (meta)data. The FAIRsharing initiative provides a curated, searchable resource to help find many of these. The disease ontology we have mentioned already in the first exercise has its own page in FAIRsharing. Another useful resource serving the data needs of specific communities is RDMkit. RDMkit is an online research data management toolkit for Life Sciences, and as part of its product hosts pages for domain-specific best practices and guidelines. Domain pages signpost detail and promote relevant considerations, tools and resources.
Exercise
Familiarise yourself with the Bioimaging data domain page on RDMkit. Read the section on “Standard (meta)data formats” about a third of the way down.
What 3 URLs are given to help a researcher gather appropriate metadata for their images?Solution
OME model XML-based representation of microscopy data.
Quality assessment working groups
REMBI
Useful resources
- More about Metadata: RDMkit, FAIR Cookbook
- Definition of Machine readability: Research Data Alliance (RDA)
- Definition of Qualified identifiers: GO FAIR
- More about Data licensing: RDMkit, FAIR Cookbook
- Definition of Metadata community standards: FAIRDOM
- Registry of FAIR metadata standards: FAIRsharing
- Making interoperable metadata: Ed-DaSH, Ten simple rules for making a vocabulary FAIR,
EOSC Guidelines for research institutes to enable discoverability of research data- Useful metadata projects and tools: Dublin Core, ISA framework, COPO
Key Points
Metadata is data about data
Metadata is used to help you find and interpret data
Domain-relevant community standards helps researchers write better, interoperable metadata
Data registration
Overview
Teaching: 20 min
Exercises: 20 minQuestions
What is data registration?
Why should you upload your data to a data repository?
What types of data repositories are there?
How to choose the right repository for your dataset?
Objectives
Describe why indexed data repositories are important
Summarise resources enabling you to choose a searchable repository
Data registration and the FAIR Principles
Data registration relates to the following 3 FAIR Principles (Table 3.1).
We will discuss and signpost these in this Episode.
| The FAIR Guiding Principles | |
|---|---|
| To be Findable: | F1. (meta)data are assigned a globally unique and persistent identifier F2. data are described with rich metadata (defined by R1 below) F3. metadata clearly and explicitly include the identifier of the data it describes F4. (meta)data are registered or indexed in a searchable resource |
| To be Accessible: | A1. (meta)data are retrievable by their identifier using a standardized communications protocol A1.1 the protocol is open, free, and universally implementable A1.2 the protocol allows for an authentication and authorization procedure, where necessary A2. metadata are accessible, even when the data are no longer available |
| To be Interoperable: | I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation. I2. (meta)data use vocabularies that follow FAIR principles I3. (meta)data include qualified references to other (meta)data |
| To be Reusable: | R1. meta(data) are richly described with a plurality of accurate and relevant attributes R1.1. (meta)data are released with a clear and accessible data usage license R1.2. (meta)data are associated with detailed provenance R1.3. (meta)data meet domain-relevant community standards |
Table 3.1: The 15 FAIR Guiding Principles. Principles relating to data registration in black.
What is data deposition and registration?
Data deposition and registration refer to the process of uploading data to a searchable resource, and providing appropriate metadata to facilitate its discoverability. For example, a data repository, where data and metadata can be uploaded, may enable it to be discovered, preserved and accessed. Here we use the general term data repository to describe any online storage location that can host deposited (meta)data.
In the context of FAIR, data deposition relates to a number of the Guiding Principles. Firstly, “(meta)data are registered or indexed in a searchable resource” (FAIR Principle F4). Searchable (indexed) metadata enables humans and computers to query and discover data of interest, though this depends on what is indexed. Here, indexing refers to a process that occurs within the architecture of the data repository (local indexing) where metadata are organised so that they can be queried based on a defined field. It is worth noting that community resources, focused on a particular domain (for example, the human database in Ensembl) are better indexed for a particular community, rather than generic repositories (for example, Zenodo) which may not index the community specific components, and may focus on higher level metadata. Indexing by an internet search engine is another example of this. Google (and other search engines, such as yahoo and yandex) have an agreed vocabulary (schema.org), within web pages, that are ‘scraped’ and indexed. While the focus of this vocabulary was originally intended for commercial products, community specific efforts to facilitate discipline-specific indexing are under way (for example, Bioschemas).
Why should I upload my data to a data repository?
Data repositories are generally preferred to file storage systems (such as Dropbox) or sharing data on an ad hoc basis, since they often better support FAIR best practice. Repositories will assign citable, “globally unique and persistent identifiers” (FAIR Principle F1) to data, and in some cases enable a data submitter to apply a data usage licence through association with the resource (FAIR Principle R1.1).
Although not exclusively, data repositories support the creation of metadata through curation interfaces providing drop-downs and text fields for metadata entry and validation. Often in the case of a domain or data-specific data repository, such as BioStudies shown in the previous Episode, drop-downs for metadata curation will link community-endorsed vocabularies (FAIR Principle R1.3).
Types of data repository
General public data repositories, such as Zenodo, are multidisciplinary and permit registration and upload of open and closed access (meta)data. Metadata curation is relatively high level and made searchable via indexing. Relating to data in the Life Sciences, Zenodo is often used to publish and provide citable URLs to supplementary data within articles, usually in instances where a domain repository does not exist.
Institutional repositories work similarly and provide an online archive for hosting, indexing and preserving research output specific to an institution. Typically these house more than data, providing a repository often for documents and articles. Institutions will have their own systems supported locally or buy into company solutions.
Discipline-specific repositories cater for communities and datatypes, and typically provide web interfaces to annotate rich metadata at the point when data are submitted. Examples of these belong to the suite of data repositories at the European Bioinformatics Institute (EBI) where rich metadata creation is supported by teams of curators.
Exercise
An example of a discipline specific repository is ArrayExpress database. ArrayExpress stores data from high-through functional genomics assays, such as RNAseq, ChIPseq and expression microarrays. The data submission interface of ArrayExpress is called Annotare. Without creating a login, what help is given to a person looking to submit a dataset for the first time?
Solution
Both a submission guide and YouTube video is provided.
Exercise
Finding more help on how to upload data to specific repositories The FAIR Cookbook is an online open resource housing specific ‘how to’ guides or recipes. Use the FAIR Cookbook to find two recipes for “depositing data to Zenodo” and “registering datasets with Wikidata”, respectively.
Solution
Open the Findability pulldown on the left hand banner to find recipes for the following: Depositing to generic repositories - Zenodo use case and Registering Datasets in Wikidata.
Exercise
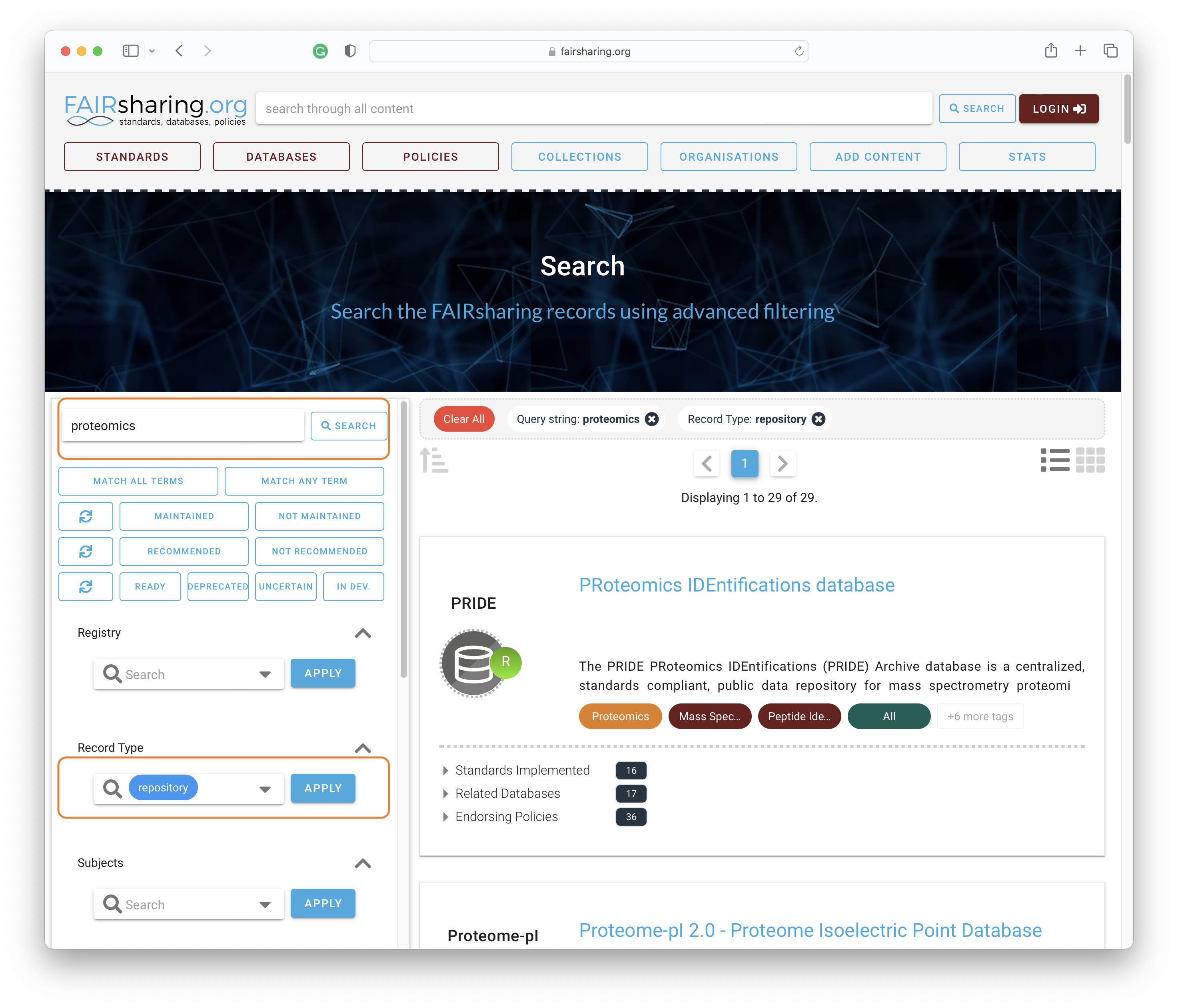
Choosing the right data repository for your data FAIRsharing helps researchers identify suitable data repositories, standards and policies relating to their data. Use this resource to identify data repositories for proteomic data.
Solution
Access the search bar for the FAIRsharing database registry. Search for proteomics and select “repository” under “Record Type”.
Useful Resources
- Registries and lists of public repositories: FAIR Cookbook and nature journal
- Publishing your data: RDMkit
- Using Bioschemas to embed metadata into webpages: FAIR Cookbook Bioschemas
Key Points
A good way to FAIRify your (meta)data is through submission to a public repository, if it indexes and exposes the appropriate level of metadata to serve your specific use case or serve your envisaged users
Use Repositories that support controlled access to data if necessary
FAIRsharing is a useful resource to locate relevant public repositories
Access
Overview
Teaching: 40 min
Exercises: 10 minQuestions
What is data access in the context of FAIR
What are the different types of data access?
What is a data usage licence?
How can you share sensitive data?
Objectives
To illustrate data access in terms of the FAIR Principles using companion terms including communications protocol and authentication
To interpret the data usage licence associated with different data sets
Data access and the FAIR Principles
Data access relates to the following 5 FAIR Principles (Table 4.1). We will discuss and signpost these in this episode.
| The FAIR Guiding Principles | |
|---|---|
| To be Findable: | F1. (meta)data are assigned a globally unique and persistent identifier F2. data are described with rich metadata (defined by R1 below) F3. metadata clearly and explicitly include the identifier of the data it describes F4. (meta)data are registered or indexed in a searchable resource |
| To be Accessible: | A1. (meta)data are retrievable by their identifier using a standardized communications protocol A1.1 the protocol is open, free, and universally implementable A1.2 the protocol allows for an authentication and authorization procedure, where necessary A2. metadata are accessible, even when the data are no longer available |
| To be Interoperable: | I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation. I2. (meta)data use vocabularies that follow FAIR principles I3. (meta)data include qualified references to other (meta)data |
| To be Reusable: | R1. meta(data) are richly described with a plurality of accurate and relevant attributes R1.1. (meta)data are released with a clear and accessible data usage license R1.2. (meta)data are associated with detailed provenance R1.3. (meta)data meet domain-relevant community standards |
Table 4.1: The 15 FAIR Guiding Principles. Principles relating to data access are highlighted in black.
What is data access?
Making data accessible means it can be made available to use by both humans and computers, though FAIR data does not necessarily mandate that all data is openly accessible and we discuss this in a minute.
As outlined in the Accessibility Principles (meta)data must be retrievable by their identifier using a standardised communication protocol (FAIR Principle A1) which is open, free and universally implementable (FAIR Principle A1.1).
A standardised communication protocol is something like http(s) or ftp that allows data to be requested and downloaded, for example, by clicking on a link on a webpage. Simply put, a protocol is a method that connects two computers and ensures secure data transfer. Web browsers such as Firefox and Chrome are able to use the http(s) communication protocol since it is universally implementable, open and free.
It is important to note that (meta)data access is not limited to humans clicking links on webpages. For a machine ‘user’, examples of accessing data include the use of an ‘application programming interface’ (API), and Unix command line, using wget and curl.
We mentioned above that FAIR data does not necessarily mandate open data. Commonly, this relates to controlled access, which is discussed later in this Episode, but in the exercise below we investigate a dataset that has been deleted where it has been previously accessible. FAIR states: “metadata are accessible even when the data are no longer available” (FAIR Principle A2) which is one of the few principles that relates solely to metadata (and not data). This means data can be deleted at any time from its original online location, but its original metadata must remain accessible. PIDs (usually connected through URLs) to the original data must remain live, though the record may, for example, change to display metadata only. This is useful where originally referenced data changes over time, or becomes obsolete or deprecated; a record of the original metadata, and where appropriate reasons for its removal, or redirection to updated records, provide a provenance trail from the original data, which may have been referenced, for example, in publications, where it is not possible to update the PID.
Exercise
The Protein Data Bank in Europe (PDBe) is a searchable repository of biological macromolecular structures. Look at the following record that has been retired: 1ins. You will see that the crystal structure is no longer available, but what metadata is available?
Solution
Metadata includes the original citation, an explanation of why the record is no longer available and a redirection to the replacement entry.
Types of data access
Where restricted access is required (for example, sensitive data or data subject to intellectual property) generally only the recommended parts are restricted, whereas associated metadata and non-sensitive data are openly accessible. As part of the FAIR Principles, terms of access need to be stated, usually as part of a data licence (FAIR Principle R1.1). Remember, FAIR data is as open as possible, and as closed as necessary. Resources for working with sensitive data are given at the end of this Episode.
There are four types of data access as described in RDMkit:
Open access: Anyone can access the data, and use it for any purpose.
Registered access or authentication procedure: researchers are required to register and authenticate to have the right to access the data (login and password)
Controlled access or Data Access Committees (DACs): researchers will apply for access, and their application reviewed by a data access committee
Access upon request (not recommended): a researcher provides their contact details for access. Contact details should be provided in the metadata which will be publicly available.
Any access requiring login and password, makes use of “[a protocol] allowing for an authentication and authorisation procedure, where necessary” (FAIR Principle A1.2). Commonly for authentication, a researcher will be assigned a unique ID, or the system may support sign-in with an ORCID ID, which is the case of many data repositories including Zenodo. In some cases there may be an option to use a google account or institutional email to sign in, and many infrastructures also support ‘single sign in’.
Data usage licence
A data usage licence describes the legal rights on how others use your data. When you publish your data, you should describe clearly in what capacity your data can be used.
There are many types of licences that can be used, including the MIT licence (for software) or the commonly used Creative Commons licences (a selectable collection of licences). These licences provide precise descriptions of the rights to data use, where the latter defines rights for sharing, adapting and commercialisation. Open access data usually carries the CC BY 4.0 or CC0 licence permitting open sharing and adaptation, even for commercial purposes. The licence is applied by adding the licence declaration to the data similarly to this training page. Take a look at the banner at the bottom of this page. It states: Licensed under CC-BY 4.0 2018–2023 by The Carpentries.
Making sensitive data accessible
Controlled access is often afforded to sensitive data or commonly any data that could potentially do harm, see RDMkit for a full definition.
Where access is granted, sensitive data is often de-identified, meaning that identifying (meta) data is removed or reassigned, leaving the analytics-appropriate component.
For example, a participant’s name can be removed from a questionnaire, and their home address can be substituted for the name of the town they live in. This anonymises the data, since the participant can no longer be located. Note though that other information in the questionnaire could compromise this. If other data reveal that the participant won the town’s 10K road race in 2023, we could potentially identify the individual using the name of the town and an online search. If more information in the questionnaire states that the participant has a rare disease, we are broaching disclosure of sensitive, personal data.
Though people refer to anonymisation when de-identifying (meta)data, often they mean pseudonymisation. Data anonymisation and pseudonymisation are slightly different.
Data anonymisation is the process of irreversibly de-identifying personal data such that an individual cannot be identified by anyone, including the study team and the individual themselves. If data are anonymised, no one can link data back to the subject.
Pseudonymisation is more commonly used. It is a process where identifying-fields are replaced by artificial identifiers called pseudonyms or pseudonymised IDs. Commonly, a person’s name or medical ID, will be replaced with a unique participant ID within the study. Pseudonymisation ensures no one can link data back to the individual, apart from nominated members of the study team who will be able to link pseudonyms to identifying information, such medical records.
Exercise
Use RDMkit’s guidelines on sensitive data to familiarise yourself further on de-identification of data. What further training can you identify?
Solution
At the bottom of the page, under “Training”, useful resources are given. The TeSS training portal permits users to search for courses, events, videos and other learning material for data in the life sciences.
Useful resources
- More about Data Access and Sharing: RDMkit, FAIR Cookbook
- Definition and examples of Standardised Communication protocol using authentication: Australian Research Data Commons
- More about Data licensing: RDMkit, FAIR Cookbook
- The Creative commons licences
- Working with sensitive data: EBI training
Key Points
Data access is supported by standardised communication protocols allowing for authentication where appropriate
Metadata must be available even if data are deleted
Data usage licences are applied to (meta)data detailing terms of use
Sensitive data can be subject to restricted access and/or de-identification
Persistent identifiers
Overview
Teaching: 40 min
Exercises: 10 minQuestions
What is a persistent identifier?
What is the structure of identifiers?
Why it is important for your dataset to have an identifiers?
Objectives
Explain the definition and importance of using identifiers
Illustrate what are the persistent identifiers
Give examples of the structure of persistent identifiers
Persistent identifiers and the FAIR Principles
Data Identifiers relate to the following 5 FAIR Principles (Table 5.1). We will discuss and signpost these in this episode.
| The FAIR Guiding Principles | |
|---|---|
| To be Findable: | F1. (meta)data are assigned a globally unique and persistent identifier F2. data are described with rich metadata (defined by R1 below) F3. metadata clearly and explicitly include the identifier of the data it describes F4. (meta)data are registered or indexed in a searchable resource |
| To be Accessible: | A1. (meta)data are retrievable by their identifier using a standardized communications protocol A1.1 the protocol is open, free, and universally implementable A1.2 the protocol allows for an authentication and authorization procedure, where necessary A2. metadata are accessible, even when the data are no longer available |
| To be Interoperable: | I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation. I2. (meta)data use vocabularies that follow FAIR principles I3. (meta)data include qualified references to other (meta)data |
| To be Reusable: | R1. meta(data) are richly described with a plurality of accurate and relevant attributes R1.1. (meta)data are released with a clear and accessible data usage license R1.2. (meta)data are associated with detailed provenance R1.3. (meta)data meet domain-relevant community standards |
Table 5.1: The 15 FAIR Guiding Principles. Principles relating to data identifiers are highlighted in black.
Using persistent identifiers (PIDs)
Identifiers are an important theme within the FAIR principles, arguably being foundational; they are considered two of the pillars for the FAIR principles, since they are crucial for Findable (F) and Accessible (A) principles.
Identifiers are an eternal reference to a digital resource such as a dataset and its metadata. They provide the information required to reliably identify, verify and locate your research data.
Commonly, a persistent identifier is a unique record ID in a database, or a unique URL that takes a researcher to the data in question in a database. Persistent identifiers (PIDs) have to be unique so that only one dataset can be identified by this identifier. In addition to the identifier being unique, it needs to be persistent. When depositing or hosting data, you should ensure the longevity of this persistence meets your requirements, which may require reading specific database policies regarding identifier policy.
Since FAIR permits withdrawal of data, the FAIR Principles combat the potential for broken URLs by stating: “Metadata are accessible, even when the data are no longer available.” (FAIR Principle A2). This means the link (PID) remains valid, displaying all the original metadata of the record even though the data is no longer available.
It is important to note that when you upload your data to a public repository, the repository will create this ID for you automatically.
Based on how to FAIR, there are many resources that can help you find databases to assign PIDs to your data. One of these resources is FAIRsharing something we’ve already encountered in the previous episodes. FAIRsharing, provides a list of databases grouped by domains and organisations.
The structure of persistent identifiers
To ensure that a PID is globally unique, commonly it is supplied as a unique URL. For the case of a record in a biological database, the use of a URL ensures that the database record ID is associated with the database name or often some derivation of this. This is often enough to ensure the uniqueness of the PID for any future scenario.
Commonly, for things like publications, a DOI is used for the PID, where DOI stands for Digital Object Identifier. An example is shown below where the PID is constructed from 3 pieces of information: the resolver service, the prefix (namespace) and the suffix (dataset ID).
Resolver service: the domain/service/institution hosting the PID e.g. [https://www.doi.org] (doi.org)
Prefix: a unique number referring to the publisher. This is also known as the namespace.
Suffix: the unique dataset number
 Figure 5.1: The structure of a DOI
Figure 5.1: The structure of a DOI
For biological data, PIDs usually require a resolver that can deal with multiple resolving locations, which means that if a database changes its name or internal structure, the new and old variations of the PID remain valid and take the user to the (meta)data. One commonly used resolver service is called identifiers.org which maintains a list of database namespaces (prefix’s) as a persistent record. If a database changes its name, it keeps the original namespace operational, or alternatively arranges for redirection from the original.
Examples of using identifiers.org to construct a PID are given below for 2 different databases, Ensembl and WikiPathways, respectively. The namespace is given as the database name in these examples.
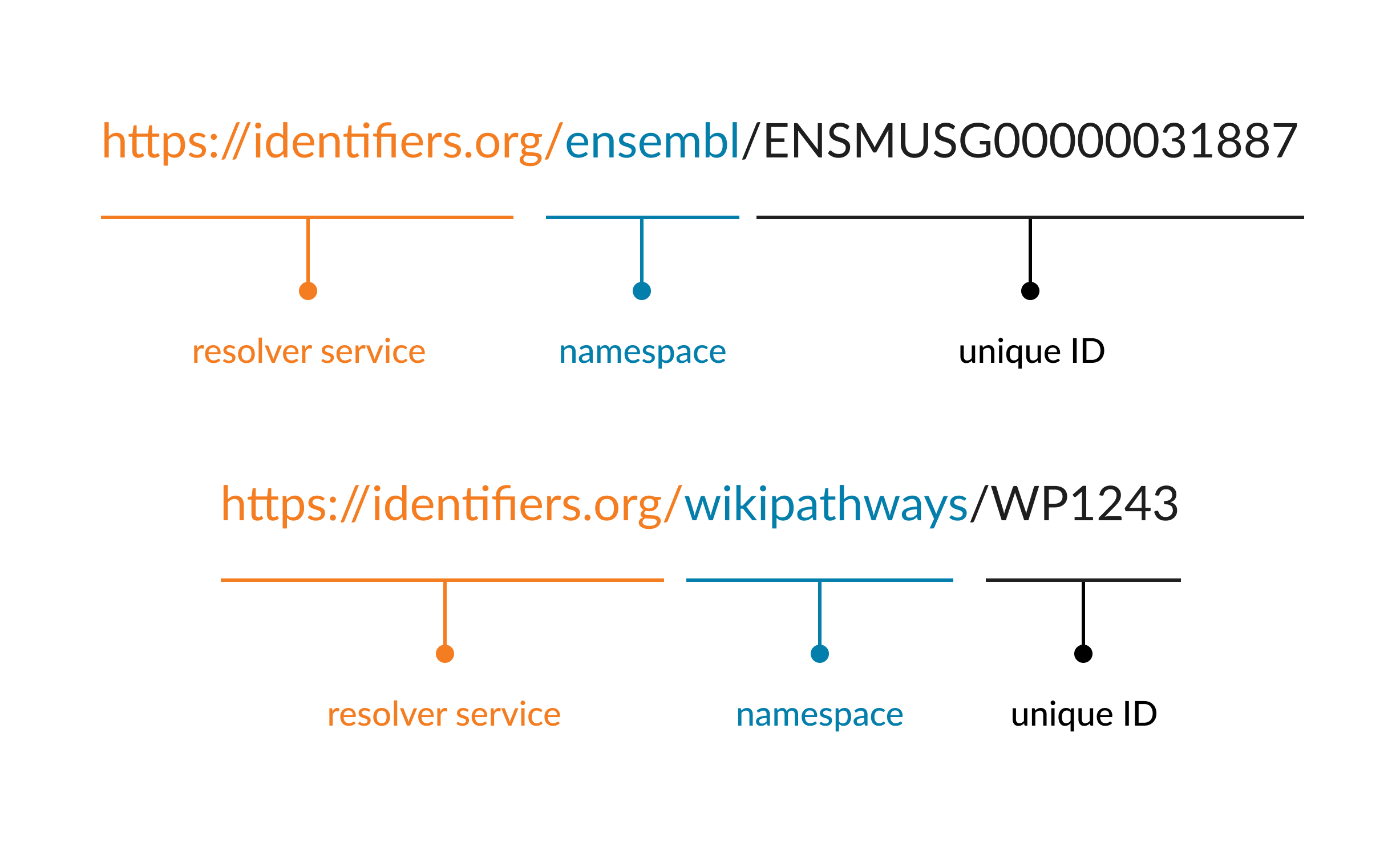 Figure 5.1: The structure of a PID using the identifiers.org resolver service
Figure 5.1: The structure of a PID using the identifiers.org resolver service
Exercise:
Access the preprint for Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. This paper makes 10 recommendations for PID best practice.
Locate the lesson for “Do not reassign or delete identifiers”. Which PID is used as an example of a “tombstone page”? Which FAIR Principle does this relate to?
Solution
Tombstone PID: https://www.uniprot.org/uniprotkb/A0AV18
This relates directly to the FAIR Principle A2. “Metadata are accessible, even when the data are no longer available” Note that other FAIR Principles are also illustrated.
The Tombstone page is “retrievable by their identifier using a standardised communications protocol”(FAIR Principle A1), which in this case is http(s). The page contains metadata which “are assigned a globally unique and persistent identifier” (FAIR Principle F1). Also the metadata “clearly and explicitly includes the identifier of the data they describe” (FAIR Principle F3) noting that the identifier itself is featured on the webpage.
Additionally, the webpage features a link to the updated UniProt record thereby “metadata include qualified reference to other (meta)data” (FAIR Principle I3)
Useful Resources
- More on identifiers: RDMkit and FAIR Cookbook
- Nick Juty, Sarala M. Wimalaratne, Stian Soiland-Reyes, John Kunze, Carole A. Goble, Tim Clark; Unique, Persistent, Resolvable: Identifiers as the Foundation of FAIR. Data Intelligence 2020; 2 (1-2): 30–39. doi: https://doi.org/10.1162/dint_a_00025
Key Points
PIDs are eternal and unique
PIDs are commonly URLs in the Life Sciences